
Existing software libraries for Topological Data Analysis (TDA) offer limited support for interactive visualization. Most libraries only allow to visualize topological descriptors (e.g., persistence diagrams), and lose the connection with the original domain of data. This makes it challenging for users to interpret the results of a TDA pipeline in an exploratory context.
TopoEmbedding is a web-based tool that simplifies the interactive visualization and analysis of persistence-based descriptors. TopoEmbedding allows non-experts in TDA to explore similarities and differences found by TDA descriptors with simple yet effective visualization techniques.
An interactive showcase of TopoEmbedding can be accessed here.
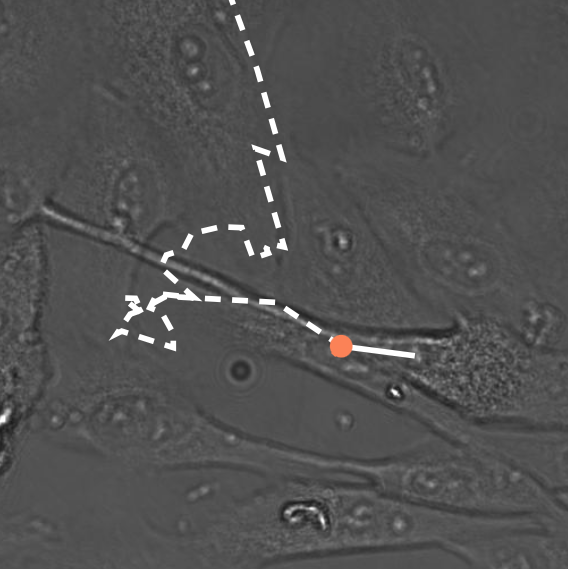
CellTrackViz allows to visualize the result of cell tracking algorithms interactively investigating their performance.
In our Eurovis paper we illustrate the importance of analyzing cell tracking algorithm performance in order to pick the best suited approach based on the biologists' need.
An interactive showcase of CellTrackViz can be accessed here.
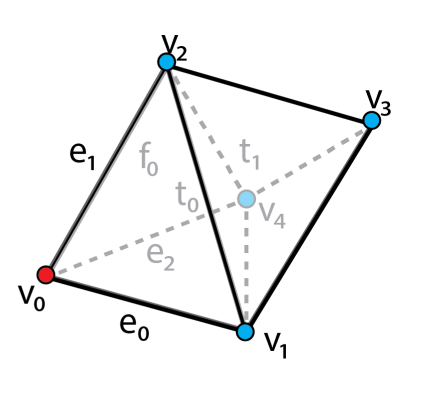
TopoCluster is a new block-based data structure for the efficient encoding and processing of irregular data. The data structure provides efficient computation of mesh connectivity with a low memory footprint.
TopoCluster can be used as a standalone C++ library, or from within the Topology ToolKit (TTK). The transparent integration allows to run any plugin of TTK using TopoCluster saving up to an order of magnitude of memory.
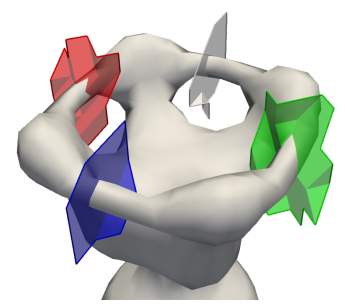
The persistence cycles toolbox implements an efficient algorithm for computing representative cycles of persistent homology.
In our associated TVCG paper we illustrate the importance of rendering persistence cycles when analyzing scalar fields, and we discuss the advantages that our approach provides compared to other techniques in topology-based visualization.
This toolbox provides an efficient implementation of our approach , as a new module for the Topology Toolkit (TTK).